Genetic Variant Classification (Classification)
For my third Metis project, I predicted whether a genetic variant is likely to be classified in a conflicting manner or not. Genetic variants are classified at labs (usually manually) in one of 5 different ways in terms of clinical significance:

These are split into 3 categories:
- pathogenic or likely pathogenic
- variant of uncertain significance
- benign or likely benign
When a variant is classified in a different category by separate labs, that classification is said to be “conflicting.” By identifying which gene variants are likely to be classified in a conflicting manner or not:
- Biologists and genetics labs can better identify those variants which require further study and lab testing
- Physicians can better plan patient treatment and respond to lab results
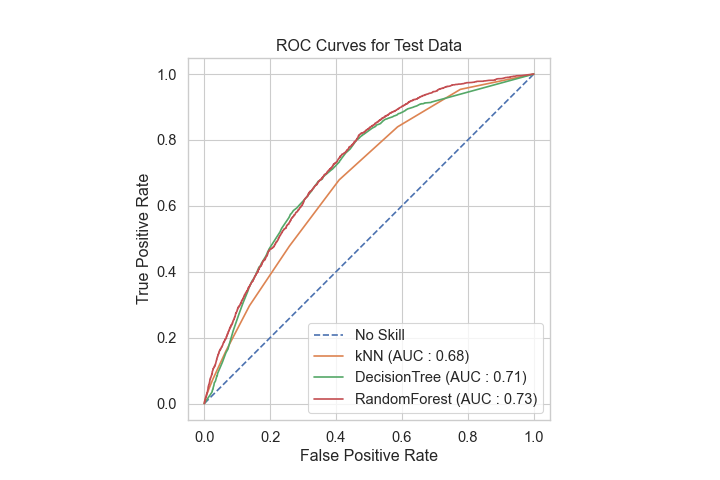
I used a Kaggle dataset for this project, and built and tested various clssification models. The figure below shows a comparison of the model ROC curves. I ultimately selected the RandomForest model since it performed the best and was still relatively interpretable. Please see the project GitHub page for more extensive details.